 
Lipid-protein interaction
Together with lipids, proteins are the main
constituents of natural membranes, and are implicated in all
biochemical processes occurring in the membrane. Membrane proteins
include integral proteins that are embedded in the bilayer or span the
membrane, and peripheral proteins that are bound to the membrane
surface. Both these types of proteins are studied in our lab. Whereas
the knowledge of the phase behavior of multicomponent lipid systems is
the starting point, the understanding of biomembranes requires
knowledge about the molecular interactions of all components, notably
lipid-protein interactions.
Relevant problems that are addressed in our
group using fluorescence methodologies are:
- Quantification of the extent of interaction of the
peptide/protein with the membrane (fluorescence
intensity/lifetime/anisotropy, Figure 1);
|
|
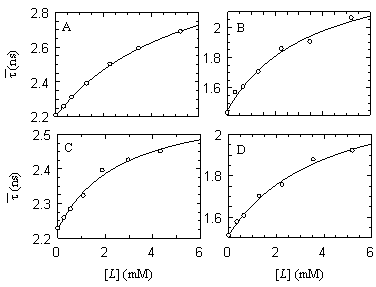
Figure 1 - Variation of the
lifetime-weighted quantum yield 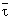 of α-MSH ( λexc = 295 nm) vs. lipid
concentration [L] of DMPC/DMPG (3:1) (A and B), and DMPC/DMPA (3:1) (C
and D) in the gel phase at 20ºC (A and C) and in the fluid phase
at 37ºC (B and D), as increasing amounts of peptide partition to
the bilayers. Peptide concentration (~30 μM) was kept constant during
the experiments. The solid line is the fit of the partition model to
the data. of α-MSH ( λexc = 295 nm) vs. lipid
concentration [L] of DMPC/DMPG (3:1) (A and B), and DMPC/DMPA (3:1) (C
and D) in the gel phase at 20ºC (A and C) and in the fluid phase
at 37ºC (B and D), as increasing amounts of peptide partition to
the bilayers. Peptide concentration (~30 μM) was kept constant during
the experiments. The solid line is the fit of the partition model to
the data.
|
- Determining the transverse location of the peptide/protein
fluorophore (quenching, FRET, spectral changes);
- Obtaining information on the secondary structure (Figure
2)/dynamics of the membrane-bound peptide/protein (fluorescence
intensity and anisotropy decays);
|
|
Figure 2 - Model of the β-hairpin
proposed for the ShB peptide inserted into palmitoyloleoylphosphatidic
acid (POPA) vesicles. Phospholipids in the bilayer are colored in
yellow. The peptide surface is colored according to the electrostatic
potential (blue, electropositive, and red, electronegative charges).
The β-structure backbone and Tyr-8 residue are colored in green. The
dotted lines represent the bilayer surface.
|
- Formation of protein/peptide-rich patches or
protein/peptide aggregates vs. random distribution (fluorescence
self-quenching, FRET (both homo- and heterotransfer, Figure 3));
|
|
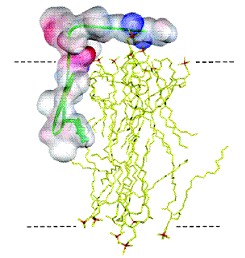
Figure 3 - Putative organization of
γM4 in Chol-poor and Chol-rich systems. A plausible structural
framework to account for the absence of energy migration between Trp453
residues in the γM4 peptide, and the variation of lifetime-weighted
quantum yield with peptide concentration on the ld and lo phases is
provided. A) Top view of the α-helical peptide showing the relative
positions of Trp453 and Cys451, and a probable
geometry for a disulfide bonded dimer. B) Possible geometry for a
linear aggregate. This structure is ruled out. C) Parallel aggregate in
end-on view. This structure is not ruled out by energy homotransfer
data. D) Anti-parallel aggregate, lateral view. This structure is not
ruled out by energy homotransfer data. E) Peptide-rich patch. The model
depicts the distribution of γM4 in POPC/Chol vesicles with high Chol
content (lo phase). The negative mismatch causes a disordering effect
in the vicinity of a peptide molecule, and other peptides accommodate
better in a region close to where other peptides localize. In the ld
(Chol-poor phase) the peptide is randomly distributed due to the very
good matching between the hydrophobic thickness of the bilayer and the
peptide.
|
- Formation of protein/peptide-rich patches or
protein/peptide aggregates vs. random distribution (fluorescence
self-quenching, FRET (both homo- and heterotransfer, Figure 3));
|
|
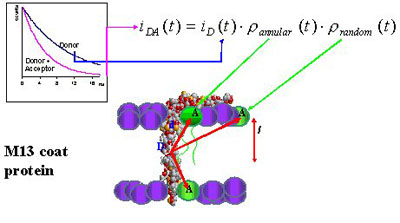
Figure 4 - Schematic representation
of M13 major coat protein in a bilayer, together with the FRET model
derived for characterization of the selectivity of this protein for
different lipid species.
|
- Peptide-mediated vesicle fusion/aggregation (FRET, Figure
5).
|
|
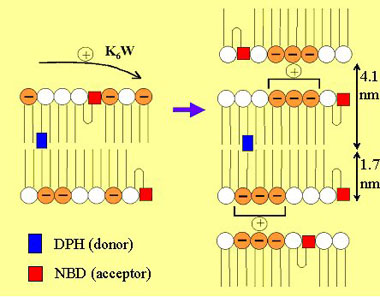
Figure 5 - Schematic representation
of the action of cationic peptide K6W upon mixtures of DPPC
(zwitterionic lipid with white headgroup in the cartoon) and DPPS
(anionic lipid with orange headgroup in the cartoon). The formation of
a multibilayer structure was unveiled by global analysis of
time-resolved FRET data.
|
go back to Research Interests
|