This study describes two Gram-negative, flexirubin-producing, biofilm-forming, motile-by-gliding and rod-shaped bacteria, isolated from the marine sponges Ircinia variabilis and Sarcotragus spinosulus collected off the coast of Algarve, Portugal. Both strains, designated Aq135T and Aq349T, were classified into the genus Aquimarina by means of 16S rRNA gene sequencing. We then performed phylogenetic, phylogenomic and biochemical analyses to determine whether these strains represent novel Aquimarina species. Whereas the closest 16S rRNA gene relatives to strain Aq135T were Aquimarina macrocephali JAMB N27T (97.8%) and Aquimarina sediminis w01T (97.1%), strain Aq349T was more closely related to Aquimarina megaterium XH134T (99.2%) and Aquimarina atlantica 22II-S11-z7T (98.1%). Both strains showed genome-wide average nucleotide identity scores below the species level cut-off (95%) with all Aquimarina type strains with publicly available genomes, including their closest relatives. Digital DNA‐DNA hybridization further suggested a novel species status for both strains since values lower than 70% hybridization level with other Aquimarina type strains were obtained. Strains Aq135T and Aq349T grew from 4 to 30°C and with between 1‐5% (w/v) NaCl in marine broth. The most abundant fatty acids were iso‐C17:03-OH and iso-C15:0 and the only respiratory quinone was MK-6. Strain Aq135T was catalase-positive and β-galactosidase-negative, while Aq349T was catalase-negative and β-galactosidase-positive. These strains hold unique sets of secondary metabolite biosynthetic gene clusters and are known to produce the peptide antibiotics aquimarins (Aq135T) and the trans-AT polyketide cuniculene (Aq349T), respectively. Based on the polyphasic approach employed in this study, we propose the novel species names Aquimarina aquimarini sp. nov. (type strain Aq135T=DSM 115833T=UCCCB 169T=ATCC TSD-360T) and Aquimarina spinulae sp. nov. (type strain Aq349T=DSM 115834T=UCCCB 170T=ATCC TSD-361T).
Aquimarina aquimarini sp. nov. and Aquimarina spinulae sp. nov., novel bacterial species with versatile natural product biosynthesis potential isolated from marine sponges
Read the full article here!
Couceiro JF, Marques M, Silva SG, Keller-Costa T, Costa R. Aquimarina aquimarini sp. nov. and Aquimarina spinulae sp. nov., novel bacterial species with versatile natural product biosynthesis potential isolated from marine sponges. Int J Syst Evol Microbiol. 2024 Jan;74(1). doi: 10.1099/ijsem.0.006228. PMID: 38240740.
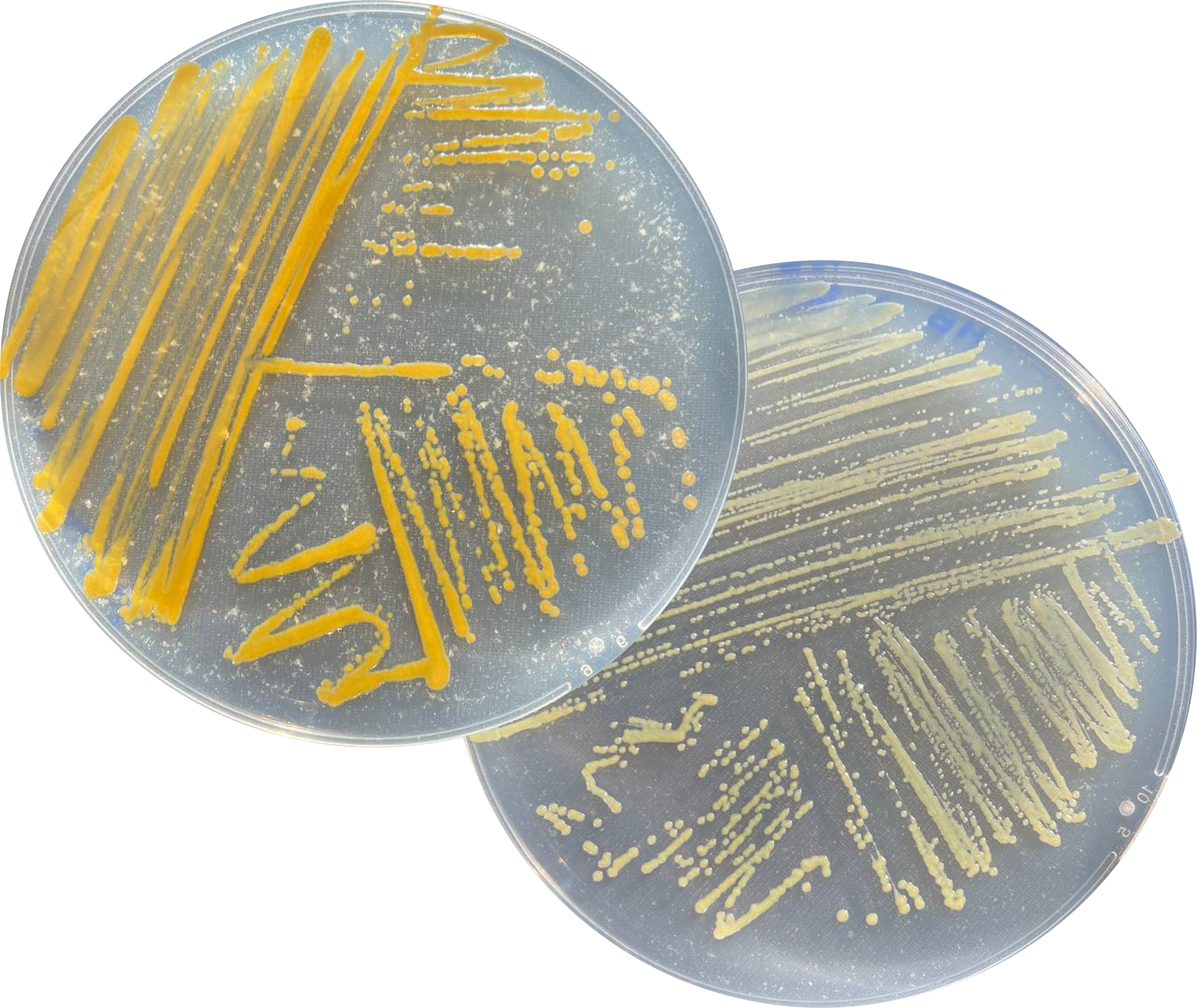
Genome Sequencing Suggests Diverse Secondary Metabolism in Coral-Associated Aquimarina megaterium
Read the full article here!
Couceiro, JF, Keller-Costa, T, Kyrpides, NC, Woyke, T, Whitman, WB, & Costa, R. Genome Sequencing suggests diverse secondary metabolism in coral-associated Aquimarina megaterium. Microbiology Resource Announcements 11(11): e00620-22 (2022). https://journals.asm.org/doi/10.1128/mra.00620-22
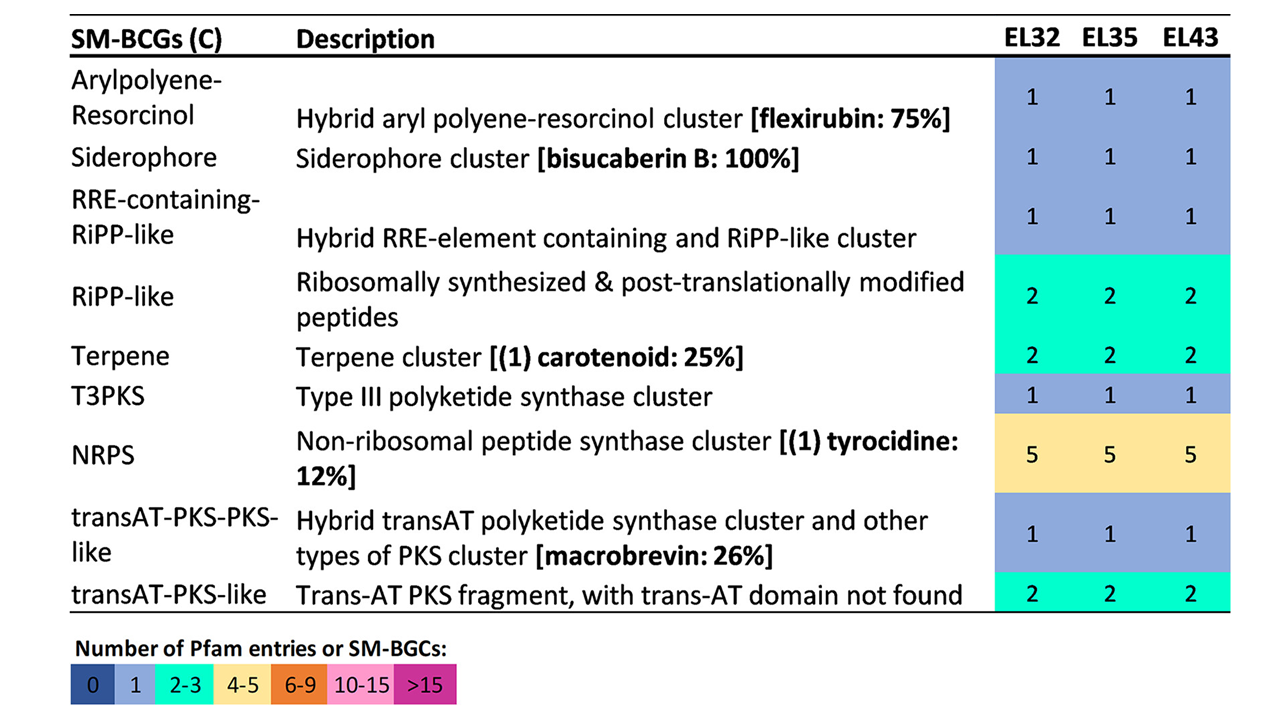
We report here the genome sequences of three Aquimarina megaterium strains isolated from the octocoral Eunicella labiata. We reveal a coding potential for versatile carbon metabolism and biosynthesis of natural products belonging to the polyketide, nonribosomal peptide, and terpene compound classes.
The Roseibium album (Labrenzia alba) Genome Possesses Multiple Symbiosis Factors Possibly Underpinning Host-Microbe Relationships in the Marine Benthos
Read the full article here!
Couceiro JF, Keller-Costa T, Marques M, Kyrpides NC, Woyke T, Whitman WB, Costa R. The Roseibium album (Labrenzia alba) Genome Possesses Multiple Symbiosis Factors Possibly Underpinning Host-Microbe Relationships in the Marine Benthos. Microbiol Resour Announc. 2021 Aug 26;10(34):e0032021. doi: 10.1128/MRA.00320-21.
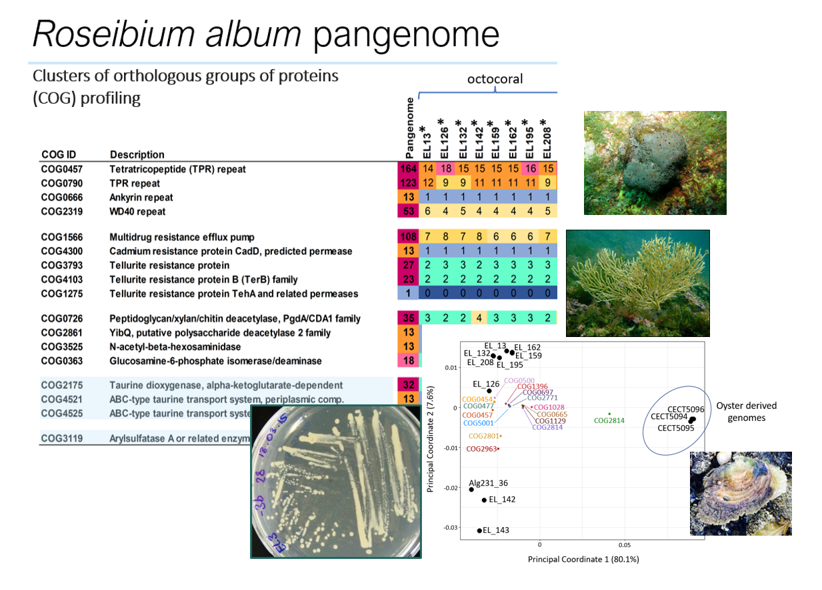
Here, we announce the genomes of eight Roseibium album (synonym Labrenzia alba) strains that were obtained from the octocoral Eunicella labiata. Genome annotation revealed multiple symbiosis factors common to all genomes, such as eukaryotic-like repeat protein- and multidrug resistance-encoding genes, which likely underpin symbiotic relationships with marine invertebrate hosts.