Suffix trees are by far the most important data structure in
stringology, with myriads of applications in fields like
bioinformatics and information retrieval. Classical representations of
suffix trees require
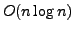
bits of space, for a string of size
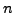
. This is considerably more than the
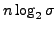
bits needed
for the string itself, where
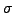
is the alphabet size. The size
of suffix trees has been a barrier to their wider adoption in
practice. Recent compressed suffix tree representations require just
the space of the compressed string plus
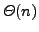
extra bits. This
is already spectacular, but still unsatisfactory when
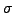
is
small as in DNA sequences.
In this paper we introduce the first compressed suffix tree
representation that breaks this linear-space barrier. Our
representation requires sublinear extra space and supports a large set
of navigational operations in logarithmic time. An essential
ingredient of our representation is the lowest common ancestor (LCA)
query. We reveal important connections between LCA queries and suffix
tree navigation.